Navigate This Page:
Genome Sequencing
Expanding the genetic and genomic resources available for shrub willow (Salix spp.) will not only facilitate improvements of the crop through breeding but will also contribute to our understanding of woody plant biology. At the forefront of this is the sequencing of the willow genome (n = 19; ~390 Mb). Larry Smart’s research group together with lead PI Dr. Gerald Tuskan and Dr. Chris Town is directing the sequencing of the genome of the female Salix purpurea L. clone 94006 at the US Department of Energy Joint Genome Institute (JGI) using next-generation sequencing technology.
To complement the next-generation sequencing of the genome, we have developed BAC and fosmid libraries in collaboration with Dr. Chris Town and Dr. Haiying Liang. Transcriptome analysis of different tissue types from S. purpurea has been completed, allowing us to characterize thousands of new genes in willow (37,864 primary and 23,656 alternative transcripts).
Expanding the genomic resources available for shrub willow (Salix spp.) will not only facilitate improvements of the crop through breeding and early selection but will also contribute to our understanding of woody plant biology.
The sequence and annotation of the female Salix purpurea clone 94006 is available at JJGI’s Phytozome Genomics Portal and the JBrowse Genome Track Browser.
Shrub Willow Genomics Group:
Genetic Mapping
The genetic basis for specific traits in shrub willow is being investigated to assist in the development of marker assisted selection for willow breeding. Molecular markers such as SNPs and SSRs as well as candidate genes linked to traits combined with phenotypic analysis are vital tools for future breeding efforts and will help predict the performance of particular varieties in certain environments and allow for major improvements to shrub willow bioenergy crops.

Among some of the characteristics of willow being investigated at the molecular level are cell wall composition and biosynthesis, phosphorus and arsenic uptake and detoxification, and the unique “corkscrew” trait observed in S. matsudana (Lin et al 2007).
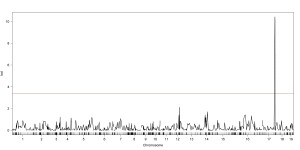
In order to make marker assisted selection successful in willow breeding, mapping populations are being produced through controlled breeding. Future research goals include the development of a linkage map for S. purpurea using SNPs from next-generation genotyping-by-sequencing (GBS) technology (Elshire et al 2011), similar to what has been used for the mapping and alignment of S. viminalis with the P. trichocarpa linkage map by Hanley et al 2006 and by Berlin et al 2010. Large F1 and F2 populations are in development for future QTL and eQTL mapping experiments.
Phylogenetics
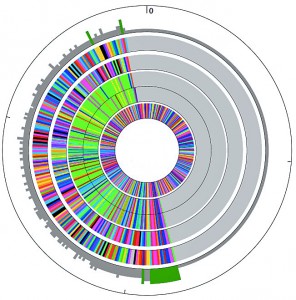
The natural dioecious sexual reproductive habit of shrub willow within large populations of overlapping generations has allowed the species to maintain high levels of genetic diversity and heterozygosity within and between populations. Thorough sampling of natural populations has provided a large gene pool of variation for potential genetic improvement. Breeding strategies for shrub willow benefit from maximizing the phenotypic variation in progeny families to increase the likelihood of identifying exceptional individuals. Research on willow breeding populations has focused on genetic diversity and heritability studies (Lin et al 2009; Cameron et al 2008; Kopp et al 2005). In addition, a complete phylogenetic analysis of important willow varieties used in our willow breeding program is now in progress using polymorphic nuclear (ITS and GBS) and plastid (matK) sequences, which will provide a better understanding of the taxonomic relationship between many of the different species and individuals currently used for breeding.
